All in your genes: new tool investigates on and off switch for genes
By Hudson Institute communications
Our genetic material can predispose us to a number of diseases and conditions. Technologies to edit or change our genes are still in their infancy, so scientists are instead looking for ways to use treatments to stop genes that cause health conditions from being turned on.
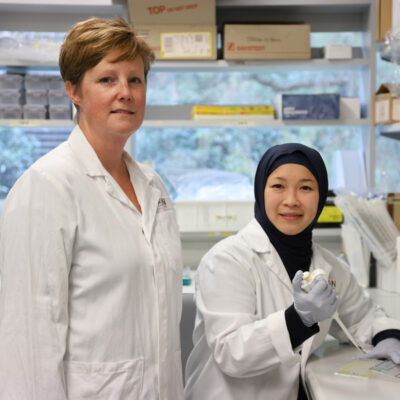
Professor Paul Hertzog and postdoctoral scientist Dr Jamie Gearing at Hudson Institute have created a computer program, CiiiDER, which predicts segments of DNA responsible for turning on and off genes. The program will be used to assist researchers in finding pathways that regulate genes.
By identifying the pathways that regulate gene expression, CiiiDER has the potential to highlight where drugs or other treatments could intervene and stop genes from being switched on. In the future, this may be applicable to a wide range of diseases, including infections, autoimmune diseases, and cancer.
There are more than 1000 proteins in humans that play a role in regulating whether a gene is turned on or not. This makes finding the protein associated with a health condition like trying to find a needle in a haystack. CiiiDER was developed as a solution to this problem.
Investigating disease
To perform a CiiiDER analysis, the user inputs a disease-associated gene signature into the program, which compares it to a normal set of DNA. CiiiDER’s analysis identifies gene regulatory proteins that are significantly over- or under-represented in the DNA of the disease gene signature, which helps identify the protein responsible for switching genes on or off.
CiiiDER is already being used by research scientists to understand gene responses to viruses such as influenza and chikungunya virus, which causes epidemics of arthritic disease.

An accessible, compatible tool
CiiiDER allows the user to analyse vast quantities of data in a matter of minutes.
Professor Paul Hertzog said the tool took several years to develop and has been made freely available online, in the interests of benefiting the wider science community, and because of its potential to pinpoint pathways that could be targeted for therapy in a wide range of conditions.
“The beauty of the program is that it can be used in combination with a variety of bioinformatics tools, such as the Interferome, to find patterns. These are tools that allow us to analyse the complexity and sophistication of the data we are presented with.”
To ensure CiiiDER is compatible with all computer operating systems, it has been written in the Java programming language.
Read about CiiiDER in the journal Plos One.
Contact us
Hudson Institute communications
t: + 61 3 8572 2697
e: communications@hudson.org.au
About Hudson Institute
Hudson Institute’ s research programs deliver in three areas of medical need – inflammation, cancer, women’s and newborn health. More
Hudson News
Get the inside view on discoveries and patient stories
“Thank you Hudson Institute researchers. Your work brings such hope to all women with ovarian cancer knowing that potentially women in the future won't have to go through what we have!”