Advanced Informatics Program

Identifying new therapeutic targets and biomarkers for low-survival paediatric cancers.
Overview
Lead Researcher | Dr Claire Sun
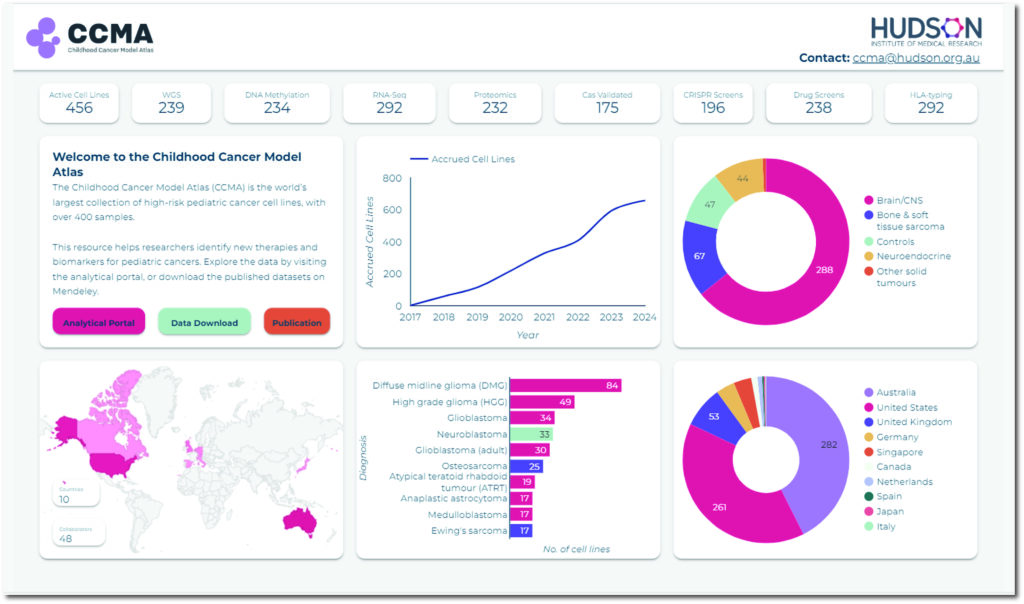
The Advanced Informatics program develops and applies state-of-the-art computational approaches to rapidly screen samples of the most challenging paediatric cancers against thousands of genetic alterations for potential therapeutic vulnerabilities. This strategy aims to identify new therapeutic targets and biomarkers for low-survival paediatric cancers. In particular, AI-based scientific methods have revolutionised multi-omics data analysis, offering an integrated view of biological systems and their complex interactions. Together, these approaches enable the prediction and prioritisation of therapeutic targets and biomarkers to advance paediatric oncology.
Multi-omics computational analysis encompasses data from
- Whole-genome sequencing (WGS)
- RNA-sequencing
- DNA methylation array
- Proteomics
- Phospho-proteomics
- Metabolomics
- Genome-scale CRISPR/Cas12 screens
- Multi-dose drug screens (n=768)
These technologies facilitate comprehensive analysis at the genome, transcriptome, epigenome and proteome levels on the Childhood Cancer Model Atlas (CCMA) models, offering a holistic view of cellular processes in paediatric cancers.
Goals
- Leverage AI-technology to identify genetic vulnerabilities, potential targeted therapies, and predictive biomarkers in paediatric cancers.
- Develop and maintain a web-based analytical data portal, providing a publicly accessible resource for visualisation and exploration of multi-omics datasets.
- Integrate patient model data analysis with clinical treatment plans for children enrolled in precision medicine programs, both nationally and internationally, to enhance clinical decision-making frameworks.
- Investigate epigenetics modifications in tumour evasion, and develop tailored immunotherapies for paediatric cancers, combining insights from epigenetics and immunology.
Advanced Informatics Program Projects
Managing the Childhood Cancer Model Atlas database

A data-driven approach to identify therapeutic targets for paediatric cancers

Targeting the dysregulated epigenome to enhance immunotherapy response